Determining Nannochloropsis cell wall and more
 Nannochloropsis’ whole cells were recently analysed using 13C-SS-NMR spectroscopy (reference n 1). Cells were grown autotrophically in a sodium [13C] bicarbonate environment and harvested during the exponential phase.
Nannochloropsis’ whole cells were recently analysed using 13C-SS-NMR spectroscopy (reference n 1). Cells were grown autotrophically in a sodium [13C] bicarbonate environment and harvested during the exponential phase.
Quantitative spectra of 13C labelled cells of Nannochloropsis oculata were analysed for resonance picks which are characteristic of carbohydrates and lipids, thus providing, among other interesting observations, useful insights on the cell wall composition of the microalga.
Need nice pics and images for your presentation?

A collection of microalgae cultures at CSIRO Marine Research. Image released by CSIRO under Creative Commons Attribution license http://scienceimage.csiro.au/tag/algae/i/1938/microalgae-collection/
The Commonwealth Scientific and Industrial Research Organization (CSIRO) has just released its Scientific Image Library under a Creative Commons Licence.
Among the many available pictures and images a small section is also dedicated to the microalgal collection of the institute. Following this link you can browse and freely download useful high quality images for lectures, reports and blog posts.
Do not forget that various collection of high quality images released under Creative Common licence already exist and some of them contain images and pictures of microalgae. For instance The Wikimedia Commons Scientific Images Collection. Don’t forget that you can easily upload your pictures and images of Nannochloropsis to the wikimedia commons collection too, as I did some time ago: https://en.wikipedia.org/wiki/File:NannochloropsisMinusNitrogenNileRed.jpg
New data on lipid accumulation in Nannochloropsis
We have already discussed in a previous post two possible models of cellular metabolism that can account for oil accumulation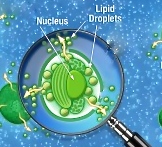 and that are in agreement with the profile of transcript and protein abundance in Nannochloropsis‘ cultures that accumulate lipids. New experimental data became recently available for the scientific community concerning this issue. Jing Li and coworkers performed a time course experiment, tracking simultaneously transcript abundance and lipid content of Nannochloropsis cultures grown in N sufficient (+N) and N depleted (-N) media. The data and their analysis were published on Plant Cell (doi: http://dx.doi.org/10.1105/tpc.113.121418). Full reference is reported at the end of this post (reference n 1).
and that are in agreement with the profile of transcript and protein abundance in Nannochloropsis‘ cultures that accumulate lipids. New experimental data became recently available for the scientific community concerning this issue. Jing Li and coworkers performed a time course experiment, tracking simultaneously transcript abundance and lipid content of Nannochloropsis cultures grown in N sufficient (+N) and N depleted (-N) media. The data and their analysis were published on Plant Cell (doi: http://dx.doi.org/10.1105/tpc.113.121418). Full reference is reported at the end of this post (reference n 1).
Families of orthologous proteins: file type
N.gaditanaB-31|Naga_100019g47.1 N.gaditanaCCMP526|Nga20827 nudix hydrolase;
Even though the differences and the imprecisions of the various gene predictions probably play a major role in the determination of the differences among the two species, a close look at the lists of proteins that are putatively assigned as characteristic of each species my reserve interesting surprises!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335.doi: 10.1093/mp/sst120
We’re looking for new authors and contributors
 Are you an expert about Nannochloropsis? Is your research focused on this organism?
Are you an expert about Nannochloropsis? Is your research focused on this organism?
Consider becoming a new author of this blog and contributing to the scientific discussion!
Why don’t you start with advertising your recent publication or commenting on the posts of this blog?
Is there any hot topic that you would like to discuss or any suggestion that you would like to ask to this scientific community?
Fill this form and we will be happy to consider you application as new author
Nannochloropsis genomes, what have we learnt?
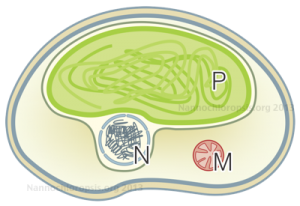
Schematic representation of Nannochloropsis cell by Andrea Telatin
Thanks to the sequencing of the genomes of two strains of Nannochloropsis gaditana [1][2] and two strains of Nannochloropsis oceanica [3][4] we now have the reference sequences to design molecular biology experiments on this microorganism. Moreover we have learnt a great deal of information, opened up new interrogatives .. and more will come analysing the data available though this portal and through the other web resources: N. gaditana CCMP526 genome and N. oceanica CCMP1779 genome.
Nannochloropsis genome is small, about 28-29 Mega bases, and very compact: there is on average 1 gene each 2.7 kilo bases in N. gaditana B-31, the genes are long on average 1.2 kilo bases and they contain very few introns.
The proteins predicted in the two species are in large part similar, about 80% of the proteins of each species are found in clusters of homologous proteins, ~60% of which accomodate proteins common to the two species. Further studies will help ruling out the limits and the inaccuracies of the gene predictions allowing to focus on the actual differences between the two species.
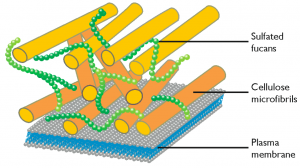
The pathways leading to the synthesis of cellulose and sulfated fucans and to the remodelling of cellulose in the cell wall have been identified both in N. gaditana and in N. oceanica, casting a light on the molecular composition of the cell wall and suggesting possible targets of genetic modification.

Recent Comments