Archive | Website
RSS for this section
FTP area updated
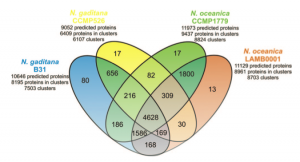
![]() We updated our FTP area, now including datasets from other Nannochloropsis species and strains and the list of families of orthologous proteins obtained from the comparison of N. gaditana and N. oceeanica predicted proteins.
We updated our FTP area, now including datasets from other Nannochloropsis species and strains and the list of families of orthologous proteins obtained from the comparison of N. gaditana and N. oceeanica predicted proteins.
Families of orthologous proteins: file type
Comparing the protein families of N. gaditana and N. oceanica, we produced the lists of exclusive proteins of each species. These lists are available for download in our FTP area.
Each file is a list of families of orthologous proteins. All the families of each file are populated only by proteins belonging to one species of Nannochloropsis. The families may contain proteins of one or more strains of the same Nannochloropsis species.
In the .txt files there is one family per line, described by the name of the organism (species and strain), then a “|”, the list of the proteins of that organism (indicated by the protein ID) the name of the following organism, another “|”, the proteins of the following organism and finally the annotation of the listed proteins.
example:
N.gaditanaB-31|Naga_100019g47.1 N.gaditanaCCMP526|Nga20827 nudix hydrolase;
Even though the differences and the imprecisions of the various gene predictions probably play a major role in the determination of the differences among the two species, a close look at the lists of proteins that are putatively assigned as characteristic of each species my reserve interesting surprises!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335.doi: 10.1093/mp/sst120
Recent Comments