Proteomics and transcriptomics findings on Nitrogen stress respons in Nannochloropsis
Nannochloropsis cultivated in normal growth condition and deprived of a nitrogen source continues growing for at least 4–5 days.
Analyses of the gene expression and of the protein abundance in nitrogen scarcity, reveal that Nannochloropsis activates mechanisms for nitrogen assimilation and redistribution to allow survival through a partial reorganisation of the cellular metabolism.
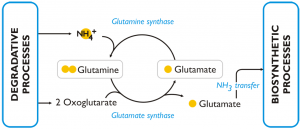
This figure reports the model of the response to nitrogen deprivation elaborated by Corteggiani Carpinelli et al. (2013).
DEGRADATIVE PROCESSES
Various genes involved in controlled degradation of proteins are over-expressed in the cells grown in nitrogen deprivation, including: cullin (Naga_100070g1), which is responsible for protein ubiquitination; ubiquitin-specific protease (Naga_100587g2); proteases (Naga_100611g3, Naga_100098g23, Naga_100015g80.1); endopeptidase Naga_101780g1); aminopeptidase (Naga_100024g51) and two autophagy-related proteins (Naga_101823g1 and Naga_100732g4), which are likely involved in the formation of cytosolic sequestering vesicles used for degradation and recycling of cellular components.
The proteomic study shows that the relative abundance of one subunit of the protein degrading complex (Proteasome subunit alpha) increases in nitrogen starvation supporting the evidence that processes of protein degradation are activated in response to nitrogen starvation. The analysis of protein abundance also suggests that in nitrogen starvation autophagy is induced. Indeed the abundance of a receptor-mediated endocytosis protein slightly increases and also that of two vacuolar proton pump subunits. These data, together with morfological observations, support the hypothesis that upon nitrogen deprivation the organelles are degraded in the vacuoles through a process of controlled autophagy.
The results of these changes in gene expression and protein abundance is the increase of degradative processes that make aminoacids available for biosynthetic processes without the need of nitrogen input (which is normally needed for the synthesis of new aminoacids).
In addition proteins involved in various degradative processes that release ammonium are also over-expressed at transcript level: guanine deaminase Naga_100022g41), which hydrolyses guanine to xanthine plus ammonia; and copper amino oxidase (Naga_100118g4), which catalyses in the two opposite directions the condensation and the release of ammonium into amines (RCH2NH2 + H2O + O2 ←→ RCHO + NH3 + H2O2).
RECYCLING
The aminoacids that are released by degradative processes can be readily incorporated into new macromolecules since the genes involved protein biosynthesis in the citosol are still transcribed in the cells during during the first 2-3 days of nitrogen deprivation. The machinery for proteins biosynthesis of the organelles are instead severely tuned down even during early nitrogen deprivation.
In nitrogen deprived cells of Nannochloropsis the GS/GOGAT pathway (L-glutamine + 2-oxoglutarate + NADPH + H+ ←→ 2 L-glutamate + NADP+) is over-expressed (both at transcript and protein level) comparing to the cells grown in nitrogen sufficient medium. The enzymes of this pathway are: glutamine synthetase (GS) (Naga_100056g25), which has a high affinity for NH3 and catalyses the incorporation of ammonia into glutamine; glutamate synthases (Naga_103301g1, Naga_101084g1, Naga_101084g2) and glutamine amidotransferase (Naga_101462g1), which are able to transfer one amino group from glutamine to 2-oxoglutarate and release, as a result, two molecules of glutamic acid. The glutamic acid produced through this pathway is an available substrate for further incorporation of intracellular ammonia and represents also a source of amine groups for cellular biosynthetic processes. The over-expression of this pathway in condition of nitrogen scarcity was observed also in other algae and in particular in diatoms.
BIOSYNTHETIC PROCESSES
As already mentioned some biosynthetic processes are still active, at least for some time, in Nannochloropsis cells after nitrogen deprivation. The balancing between selective degradation and controlled expression of genes allows the cells to reorganise and survive. Among the most considerable effects of this reorganisation there is the deescalation of the organelles and of their activity and the maintenance of the main cellular metabolism even in nitrogen shortage.
Even if a direct comparison between the transcriptomic and proteomic data is difficult due to the differences in the culturing conditions of the Nannochloropsis cells examined in the two cases, it is interesting to notice that many of the observation are actually confirmed in the two experiments, allowing to outline the main characteristics of the cellular response to nitrogen deprivation.
A summary of the informations obtained studying Nannochloropsis response to nitrogen deprivation is also reported on Wikipedia!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120
- Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR (2013) Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiology, 162:1110–1126
Tags: metacyc, nannochloropsis rna-seq, nitrogen deprivation, nitrogen recycling, proteomics, survival in nitrogen scarcity, transcriptomics, wikipedia
Recent Comments