 Nannochloropsis’ whole cells were recently analysed using 13C-SS-NMR spectroscopy (reference n 1). Cells were grown autotrophically in a sodium [13C] bicarbonate environment and harvested during the exponential phase.
Nannochloropsis’ whole cells were recently analysed using 13C-SS-NMR spectroscopy (reference n 1). Cells were grown autotrophically in a sodium [13C] bicarbonate environment and harvested during the exponential phase.
Quantitative spectra of 13C labelled cells of Nannochloropsis oculata were analysed for resonance picks which are characteristic of carbohydrates and lipids, thus providing, among other interesting observations, useful insights on the cell wall composition of the microalga.
Carbohydrate resonances in microalgae are expected from the cell walls, the headgroup of galactolipids and the storage polysaccharides.
Cell wall
Analysis of the RINEPT and CP spectra of N. oculata allowed to detect resonances in which chemical shifts agreed with those reported for cellulose. The cellulose anomeric carbon C1 only appeared in the rigid (CP) spectrum, the remaining carbons showed up in the spectra of both rigid and mobile zones. The cellulose was attributed to the cell wall (reference n 1). Analysis of the genes present in Nannochloropsis genome also showed the presence of a full set of genes that can be attributed to pathways leading to synthesis and modification of cellulose (reference n 2).
Storage sugars
Chemical shifts corresponding to both β-(1 → 3) and β-(1 → 6) glycans were detected and assigned to chrysolaminarin. Also in this case some of the resonances were exclusively detected in the CP spectra but the remaining resonances appeared on both rigid and mobile zones. The relative mobility observed could be compatible with the accumulation of chrysolaminarins in solution inside the vacuoles (reference n 1). In this case again genes associated to the biosynthesis of chrysolaminarins are found in the genome (reference n 2).
Lipids
Quantitative and qualitative analysis of the lipid content of various Nannochloropsis cultures based on MASS-spectrometry were previously reported (references n 3 and n 4). In this recent work by Arnold and coworkers (reference n 1) lipid signals were analysed from 13C-SS-NMR spectra of whole cells grown photoautotrophically.
The peak assignment showed the presence of galactosyldiacylglycerols. Peaks characteristic of the sulfolipid sulfoquinovosyl-diacylglycerol (SQDG) sugar head group seemed present on both the rigid (CP) and dynamic (RINEPT) spectra. The sugar moieties were better evidenced on the RINEPT spectrum while the glycerol moieties appeared better in the CP spectrum. Phosphatidylcholine (PC) was the dominant phospholipid detected and assigned to the plasma membrane.
The CP and RINEPT spectra were dominated by lipids’ methylene signal (30–33 ppm) consistent with a high content in saturated and mono-unsaturated fatty acid chains. The RINEPT spectrum also showed a strong signal at 128–130 ppm consistent with a high proportion of PUFAs, and possibly to EPA from the vacuolar and mobile TAGs.
For further reading on the subject I suggest of course to read the paper
References
- Arnold AA, Genard B, Zito F, Tremblay R, Warschawski DE, Marcotte I. “Identification of lipid and saccharide constituents of whole microalgal cells by 13C solid-state NMR.” Biochimica et Biophysica Acta (BBA) – Biomembranes. (2014) doi:10.1016/j.bbamem.2014.07.017
- Corteggiani Carpinelli E, Telatin A, Vitulo N, Forcato C, D’Angelo M, Schiavon R, Vezzi A, Giacometti GM, Morosinotto T, Valle G. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120.
- Li J, Han D, Wang D, Ning K, Jia J, Wei L, Jing X, Huang S, Chen J, Li Y, Hu Q, Xu J “Choreography of Transcriptomes and Lipidomes of Nannochloropsis Reveals the Mechanisms of Oil Synthesis in Microalgae.” The Plant Cell April 2014. doi: http://dx.doi.org/10.1105/tpc.113.121418
- Simionato D, Block MA, La Rocca N, Jouhet J, Maréchal E, Finazzi G, Morosinotto T. (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganisation of the photosynthetic apparatus. Eukaryotic Cell May 2013 vol. 12 no. 5 665-676

A collection of microalgae cultures at CSIRO Marine Research. Image released by CSIRO under Creative Commons Attribution license http://scienceimage.csiro.au/tag/algae/i/1938/microalgae-collection/
The Commonwealth Scientific and Industrial Research Organization (CSIRO) has just released its Scientific Image Library under a Creative Commons Licence.
Among the many available pictures and images a small section is also dedicated to the microalgal collection of the institute. Following this link you can browse and freely download useful high quality images for lectures, reports and blog posts.
Do not forget that various collection of high quality images released under Creative Common licence already exist and some of them contain images and pictures of microalgae. For instance The Wikimedia Commons Scientific Images Collection. Don’t forget that you can easily upload your pictures and images of Nannochloropsis to the wikimedia commons collection too, as I did some time ago: https://en.wikipedia.org/wiki/File:NannochloropsisMinusNitrogenNileRed.jpg
]]>
We have already discussed in a previous post two possible models of cellular metabolism that can account for oil accumulation and that are in agreement with the profile of transcript and protein abundance in Nannochloropsis‘ cultures that accumulate lipids. New experimental data became recently available for the scientific community concerning this issue. Jing Li and coworkers performed a time course experiment, tracking simultaneously transcript abundance and lipid content of Nannochloropsis cultures grown in N sufficient (+N) and N depleted (-N) media. The data and their analysis were published on Plant Cell (doi: http://dx.doi.org/10.1105/tpc.113.121418). Full reference is reported at the end of this post (reference n 1).
and that are in agreement with the profile of transcript and protein abundance in Nannochloropsis‘ cultures that accumulate lipids. New experimental data became recently available for the scientific community concerning this issue. Jing Li and coworkers performed a time course experiment, tracking simultaneously transcript abundance and lipid content of Nannochloropsis cultures grown in N sufficient (+N) and N depleted (-N) media. The data and their analysis were published on Plant Cell (doi: http://dx.doi.org/10.1105/tpc.113.121418). Full reference is reported at the end of this post (reference n 1).
The glycerolipid profiling performed by Li and coworkers revealed small amounts of TAGs in cells of N. oculata IMET1 grown in Nitrogen sufficient media and the amount of TAGs was found to increase by about 3 folds when the cultures reached the end of the lag phase. In Nitrogen depleted cultures the amount of TAGs increased by approximately 400 folds in 96 hours after Nitrogen depletion and at the same time photosynthetic membrane lipids decreased significantly. Lipase-mediated membrane degradation is likely to channel FA from photosynthetic membranes to TAGs in Nitrogen depletion and indeed 14 of the 34 putative lipases identified in the genome, were up-regulated to some extent in Nitrogen scarcity. As previously observed by Simionato et al. 2013, however, the decrease of membrane lipids is not enough to account for the increase of TAGs and de novo biosynthesis of FA must take place in the cells in Nitrogen scarcity.
According to the new experimental data, the authors hypothesise that most of the proteins involved in de novo synthesis of FA (ACCase; ACP; KAS) are always abundant in the cells, both in +N and in -N conditions, and that the increase in FA must be rather due to an increased flux of precursors that enter the FA‘s biosynthetic pathway. Previous data led other authors to drive similar conclusions (Radakovits et al. 2012; Corteggiani Carpinelli et al. 2013)
Thanks to the new experimental evidence the author argue that the data suggest the presence of multiple sub-cellular compartments involved in FA and TAG biosynthesis and that the vast majority of the newly synthesised FAs are produced in the plastid through the de novo prokaryotic pathway.
The global transcriptomic analysis suggested that various pathways, which are up-regulated in -N, could supply carbon precursors for FA synthesis during Nitrogen starvation:
- the cytosolic glycolysis pathway, which produces pyruvate
- the PDHC bypass, which yields additional acetyl-CoA
- the coupling of TCA reactions with mitochondrial b-oxidation.
The authors observe that the increase of transcripts involved in carbohydrate and protein degradation along with the identification of genes encoding plastid and mitochondrial membrane transporters suggested a shunting of carbon precursors from carbohydrate and protein catabolism into FA synthesis and ultimately TAG synthesis under nitrogen deprivation.
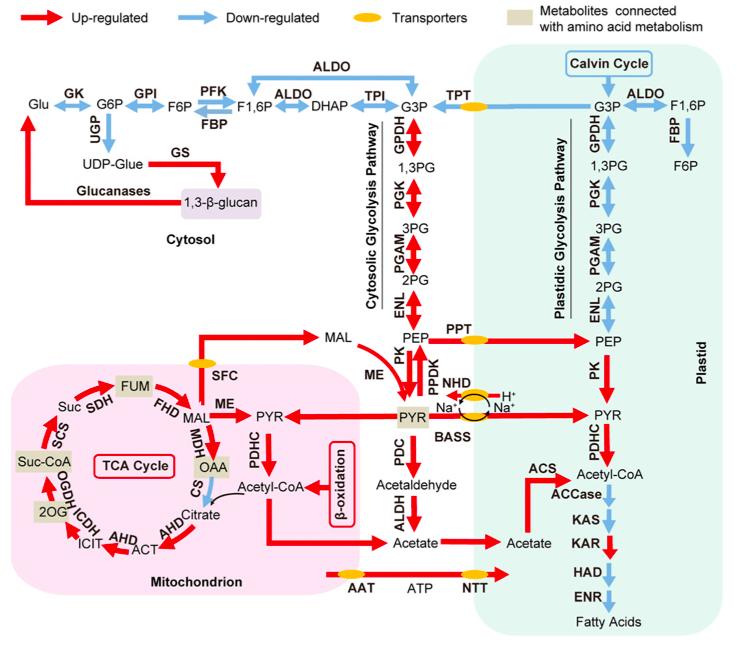
Schematic representation of the Central Carbon Metabolism in N. oculata in Nitrogen depletion from Li J et al. 2014
Finally, however, the authors suggest that, despite the increased production of FA during Nitrogen starvation, the regulatory step that determines the final TAG concentration in -N is actually the TAG assembly pathway. In oder words, under N depletion, it is the up-regulation of the transcript level of the key genes in TAG assembly, rather than those in FA biosynthesis, that leads to accelerated TAG production. Moreover the progressive up-regulation of various DGAT in Nitrogen depletion is likely very important for the bulk of the TAGs formed in -N.
I hope that this post will prompt an active debate on the topic!
References
- Li J, Han D, Wang D, Ning K, Jia J, Wei L, Jing X, Huang S, Chen J, Li Y, Hu Q, Xu J “Choreography of Transcriptomes and Lipidomes of Nannochloropsis Reveals the Mechanisms of Oil Synthesis in Microalgae.”The Plant Cell April 2014. doi: http://dx.doi.org/10.1105/tpc.113.121418
- Simionato D, Block MA, La Rocca N, Jouhet J, Maréchal E, Finazzi G, Morosinotto T. (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganisation of the photosynthetic apparatus. Eukaryotic Cell May 2013 vol. 12 no. 5 665-676
- Radakovits, R. et al. “Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana.” Nat Comms 3, 686 (2012).
- Corteggiani Carpinelli E, Telatin A, Vitulo N, Forcato C, D’Angelo M, Schiavon R, Vezzi A, Giacometti GM, Morosinotto T, Valle G. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120.
- Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR (2013) Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiology, 162:1110–1126

Confocal microscopy image of Nannochloropsis cell grown in nitrogen deprivation. Lipid droplets are in yellow and chloroplast is in red. From “Going ultra deep to unravel the secret recipe of biofuel” by Elisa Corteggiani Carpinelli
What is the mechanism by which Nannochloropsis cells synthesise and accumulate lipids in certain culturing conditions? And moreover is there a key control that we can manipulate to improve lipid yields and overcome the tradeoff between lipid production and growth? This is probably the “holy grail” of Nannochloropsis research. The general feeling is that we do not have an answer yet, but at least we have some clues to start our quest.
Transcriptomic data on Nannochloropsis gene expression during normal growth and nitrogen starvation (which leads to an evident accumulation of oil) were produced by Radakovits et al. (2012), Viler et al. (2012), Liang et al. (2012), Corteggiani Carpinelli at al. (2013). The all of this data show that genes involved in fatty acid and triacylglycerol biosynthesis are alway abundant in the cells (both when nitrogen is abundant and when it is deficient) and their expression is not correlated with the amount of oil accumulated in the cells. Also the expression of the genes involved in lipid degradation does not seem to change drastically, at least not enough to justify the effects. The general conclusion suggested by these experiments is that Nannochloropsis constitutively produces lipids and that the metabolic reorganisation that follows nitrogen deprivation increases the flux of substrates through this pathway, which is in turn capable to sustain the increased metabolic flux.
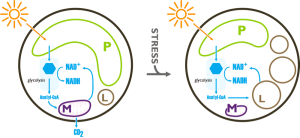
Using the available experimental evidence Corteggiani Carpinelli and coworkers (2013) propose a model of the metabolic response of Nannochloropsis to nitrogen deficiency that can lead to overproduction of fatty acids and as a consequence to the accumulation of oil. They remark that one of the most evident effects of nitrogen starvation is the deescalation of the organelles and therefore it must be taken into account in the elaboration of a model of the metabolic reorganisation of Nannochloropsis. Simionato et al. (2012) showed that despite the decrease of photosynthetic yield, photosynthesis is the only energy supply of the cells in nitrogen starvation. The model therefore states that the energy comes into the system through the photosynthetic activity of the chloroplast in both nitrogen sufficient and nitrogen depleted cultures, and produces glucose. After degradation of the glucose by glycolysis (the genes of this pathway are expressed at similar levels in both the two culturing conditions) the Acetyl-CoA and and the reduced NAD(P)H are mainly reoxidised through the Krebs cycle and the mitochondrial respiration in nitrogen sufficient cultures. Only a small fraction of the Acetyl-CoA and and the reduced NAD(P)H enter the fatty acid biosynthetic pathway in this condition. During nitrogen starvation, due to the severe down-regulation of the mitochondrial genes the reoxidation of the Acetyl-CoA and and the reduced NAD(P)H becomes less efficient and more substrates are available to enter the fatty acid biosynthetic pathway, leading to the accumulation of oil into the lipid droplets.
Potential Model of the flux trough the fatty acid synthesis pathways in nitrogen sufficient and nitrogen deficient culturing conditions based on transcriptomic data, elaborated by Corteggiani Carpinelli and coworkers (2013), drawn by Andrea Telatin. P, plastid; M, mitochondrion; L, lipid droplet.
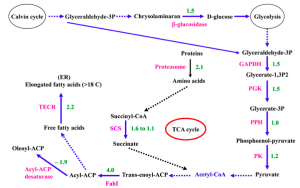
Dong et al. (2013) also examined Nannochloropsis response to nitrogen deficiency and compared protein abundance in parallele cultures grown in nitrogen sufficient and nitrogen deficient media. The experimental conditions of this experiment are different from those used in the various transcriptomic surveys: first of all the cells were supplied with CO2 during growth and moreover the time-course of the nitrogen deprivation experiment was different. Nevertheless also in this proteomic study the data seems to support the hypothesis that the accumulation of triacylglycerols is due to an increase of the metabolic flux trough the fatty acid biosynthetic pathway. The authors report that several key enzymes involved in glycolysis increase during nitrate depletion as well as enzymes involved in fatty acids synthesis and in chrysolaminaran hydrolysation. They therefore advance the hypothesis that, in their experimental conditions, the degradation of storage sugars and the up-regulation of glycolysis are responsible for the increase of substrates through the pathway.
Potential model of fatty acid synthesis pathways based on protein changes during chronic nitrogen depletion, elaborated by Dong and coworkers (2013). SCS, Succinyl-CoA synthetase; TECR, trans-2-enoyl-CoA reductase. The numbers represent fold change of proteins in relative abundance. Dashed lines mean that no related proteins were detected in this pathway.
A summary of the informations obtained studying the accumulation of lipids in Nannochloropsis in response to nitrogen deprivation is also reported on Wikipedia!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120.
- Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR (2013) Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiology, 162:1110–1126
- Liang, C. et al. “De Novo Sequencing and Global Transcriptome Analysis of Nannochloropsis sp. (Eustigmatophyceae) Following Nitrogen Starvation.” Bioenerg. Res. 6, 494–505 (2012).
- Radakovits, R. et al. “Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana.” Nat Comms 3, 686 (2012).
- Simionato D, Block MA, La Rocca N, Jouhet J, Maréchal E, Finazzi G, Morosinotto T. (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganisation of the photosynthetic apparatus. Eukaryotic Cell May 2013 vol. 12 no. 5 665-676
- Vieler, A. et al. “Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga Nannochloropsis oceanica CCMP1779.” PLoS Genet 8, e1003064 (2012).
N.gaditanaB-31|Naga_100019g47.1 N.gaditanaCCMP526|Nga20827 nudix hydrolase;
Even though the differences and the imprecisions of the various gene predictions probably play a major role in the determination of the differences among the two species, a close look at the lists of proteins that are putatively assigned as characteristic of each species my reserve interesting surprises!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335.doi: 10.1093/mp/sst120
 Are you an expert about Nannochloropsis? Is your research focused on this organism?
Are you an expert about Nannochloropsis? Is your research focused on this organism?
Consider becoming a new author of this blog and contributing to the scientific discussion!
Why don’t you start with advertising your recent publication or commenting on the posts of this blog?
Is there any hot topic that you would like to discuss or any suggestion that you would like to ask to this scientific community?
Fill this form and we will be happy to consider you application as new author
[contact-form to=’[email protected]%26#x002c; [email protected]’ subject=’application for becoming a new author of Nannochloropsis blog’][contact-field label=’Name’ type=’name’ required=’1’/][contact-field label=’Email’ type=’email’ required=’1’/][contact-field label=’Position / Title ‘ type=’url’/][contact-field label=’Expertise or Research line’ type=’textarea’ required=’1’/][/contact-form]
follow us on researchgate!
]]>Nannochloropsis cultivated in normal growth condition and deprived of a nitrogen source continues growing for at least 4–5 days.
Analyses of the gene expression and of the protein abundance in nitrogen scarcity, reveal that Nannochloropsis activates mechanisms for nitrogen assimilation and redistribution to allow survival through a partial reorganisation of the cellular metabolism.
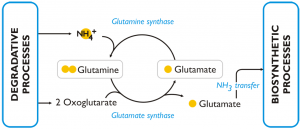
This figure reports the model of the response to nitrogen deprivation elaborated by Corteggiani Carpinelli et al. (2013).
DEGRADATIVE PROCESSES
Various genes involved in controlled degradation of proteins are over-expressed in the cells grown in nitrogen deprivation, including: cullin (Naga_100070g1), which is responsible for protein ubiquitination; ubiquitin-specific protease (Naga_100587g2); proteases (Naga_100611g3, Naga_100098g23, Naga_100015g80.1); endopeptidase Naga_101780g1); aminopeptidase (Naga_100024g51) and two autophagy-related proteins (Naga_101823g1 and Naga_100732g4), which are likely involved in the formation of cytosolic sequestering vesicles used for degradation and recycling of cellular components.
The proteomic study shows that the relative abundance of one subunit of the protein degrading complex (Proteasome subunit alpha) increases in nitrogen starvation supporting the evidence that processes of protein degradation are activated in response to nitrogen starvation. The analysis of protein abundance also suggests that in nitrogen starvation autophagy is induced. Indeed the abundance of a receptor-mediated endocytosis protein slightly increases and also that of two vacuolar proton pump subunits. These data, together with morfological observations, support the hypothesis that upon nitrogen deprivation the organelles are degraded in the vacuoles through a process of controlled autophagy.
The results of these changes in gene expression and protein abundance is the increase of degradative processes that make aminoacids available for biosynthetic processes without the need of nitrogen input (which is normally needed for the synthesis of new aminoacids).
In addition proteins involved in various degradative processes that release ammonium are also over-expressed at transcript level: guanine deaminase Naga_100022g41), which hydrolyses guanine to xanthine plus ammonia; and copper amino oxidase (Naga_100118g4), which catalyses in the two opposite directions the condensation and the release of ammonium into amines (RCH2NH2 + H2O + O2 ←→ RCHO + NH3 + H2O2).
RECYCLING
The aminoacids that are released by degradative processes can be readily incorporated into new macromolecules since the genes involved protein biosynthesis in the citosol are still transcribed in the cells during during the first 2-3 days of nitrogen deprivation. The machinery for proteins biosynthesis of the organelles are instead severely tuned down even during early nitrogen deprivation.
In nitrogen deprived cells of Nannochloropsis the GS/GOGAT pathway (L-glutamine + 2-oxoglutarate + NADPH + H+ ←→ 2 L-glutamate + NADP+) is over-expressed (both at transcript and protein level) comparing to the cells grown in nitrogen sufficient medium. The enzymes of this pathway are: glutamine synthetase (GS) (Naga_100056g25), which has a high affinity for NH3 and catalyses the incorporation of ammonia into glutamine; glutamate synthases (Naga_103301g1, Naga_101084g1, Naga_101084g2) and glutamine amidotransferase (Naga_101462g1), which are able to transfer one amino group from glutamine to 2-oxoglutarate and release, as a result, two molecules of glutamic acid. The glutamic acid produced through this pathway is an available substrate for further incorporation of intracellular ammonia and represents also a source of amine groups for cellular biosynthetic processes. The over-expression of this pathway in condition of nitrogen scarcity was observed also in other algae and in particular in diatoms.
BIOSYNTHETIC PROCESSES
As already mentioned some biosynthetic processes are still active, at least for some time, in Nannochloropsis cells after nitrogen deprivation. The balancing between selective degradation and controlled expression of genes allows the cells to reorganise and survive. Among the most considerable effects of this reorganisation there is the deescalation of the organelles and of their activity and the maintenance of the main cellular metabolism even in nitrogen shortage.
Even if a direct comparison between the transcriptomic and proteomic data is difficult due to the differences in the culturing conditions of the Nannochloropsis cells examined in the two cases, it is interesting to notice that many of the observation are actually confirmed in the two experiments, allowing to outline the main characteristics of the cellular response to nitrogen deprivation.
A summary of the informations obtained studying Nannochloropsis response to nitrogen deprivation is also reported on Wikipedia!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120
- Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR (2013) Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiology, 162:1110–1126
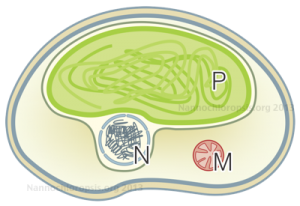
Schematic representation of Nannochloropsis cell by Andrea Telatin
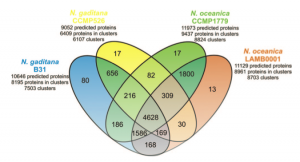
Thanks to the sequencing of the genomes of two strains of Nannochloropsis gaditana [1][2] and two strains of Nannochloropsis oceanica [3][4] we now have the reference sequences to design molecular biology experiments on this microorganism. Moreover we have learnt a great deal of information, opened up new interrogatives .. and more will come analysing the data available though this portal and through the other web resources: N. gaditana CCMP526 genome and N. oceanica CCMP1779 genome.
Nannochloropsis genome is small, about 28-29 Mega bases, and very compact: there is on average 1 gene each 2.7 kilo bases in N. gaditana B-31, the genes are long on average 1.2 kilo bases and they contain very few introns.
The proteins predicted in the two species are in large part similar, about 80% of the proteins of each species are found in clusters of homologous proteins, ~60% of which accomodate proteins common to the two species. Further studies will help ruling out the limits and the inaccuracies of the gene predictions allowing to focus on the actual differences between the two species.
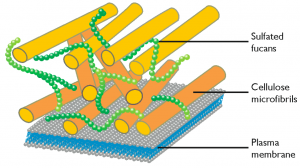
The pathways leading to the synthesis of cellulose and sulfated fucans and to the remodelling of cellulose in the cell wall have been identified both in N. gaditana and in N. oceanica, casting a light on the molecular composition of the cell wall and suggesting possible targets of genetic modification.
Moreover Nannochloropsis was shown to be able to store energy and carbon in polymers of β-1,3- and β-1,6-linked glucose called chrysolaminarins, which are very common storage sugars among the neighbouring species of microalgae. Since the genes involved in this path have been identified, they’re expression has been studied in order to better understand the metabolic fluxes in Nannochloropsis in different growth conditions.
Due to the exemplary lipid production of Nannochloropsis cultures, the genes related to lipid biosynthesis and degradation have attracted a lot of attention. A comparative analysis of Nannochloropsis genes in relation to the genes of various other algae of the red, brown and green groups has shown that Nannochloropsis has an expanded repertoire of some of the genes involved in TAG assembly. Most interestingly a high number of tag lipases (52 in N. oceanica) was found in Nannochloropsis. If you look at the information provided for each gene in this portal you will find that many of the predicted TAG lipases belong to a gene family which is exclusive to Nannochloropsis. TAG lipases can effect lipid metabolism in many ways through TAG degradation and lipid remodelling. Also in this case further studies will probably enlighten the function of these genes are their rule in Nannochloropsis metabolism.
Analysis of the genomic data of Nannochloropsis suggested that in both the two species regulatory RNAs are present and in particular miRNA. We also found genes essential for the RNA silencing process. These findings open interesting perspectives for studying Nannochloropsis using gene knock-down techniques based on the transient expression of siRNA.
Among the puzzling questions that remain open there are the capability of Nannochloropsis to reproduce sexually, the presence of a circadian regulation and the existence of motile structures in some steps of its life cycle.
While evidence of sexual reproduction has never been observed in Nannochloropsis, we found a set of genes important for meiosis and DNA recombination that are not only conserved but also actively expressed.
While shooting pictures of Nannochloropsis lipid droplets at the confocal microscopy, we found evidence of the presence of a red eye-spot in the cells, located outside of the chloroplast. We dont’t know weather this structure could be assigned as a blu-light sensor, nevertheless proteins which are orthologs of known blue light sensors were found in the genomes of Nannochloropsis, opening up the perspective of the presence of a circadian regulation of the growth and metabolic activity of this alga. Such a possibility would not be in contrast with some observations collected in outdoor cultivation plants.
Finally we found in the genome a set of proteins, which were highly similar to proteins that have been experimentally assigned to motile structures in Chlamydomonas. Also in these case the predicted genes were expressed in our experimental conditions. Evidence is definitively not enough to drive any conclusion, but certainly it rises a motivated curiosity about the possible function of these proteins..
A summary of the informations obtained after the sequencing of Nannochloropsis genomes is also reported on Wikipedia!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120
- Radakovits, R. et al. “Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana.” Nat Comms 3, 686 (2012).
- Vieler, A. et al. “Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga Nannochloropsis oceanica CCMP1779.” PLoS Genet 8, e1003064 (2012).
- Pan, K. et al.”Nuclear monoploidy and sexual propagation of Nannochloropsis oceanica (Eustigmatophyceae) as revealed by its genome sequence.” Journal of Phycology (2011), 47(6), pp.1425–1432.