New data on lipid accumulation in Nannochloropsis
We have already discussed in a previous post two possible models of cellular metabolism that can account for oil accumulation and that are in agreement with the profile of transcript and protein abundance in Nannochloropsis‘ cultures that accumulate lipids. New experimental data became recently available for the scientific community concerning this issue. Jing Li and coworkers performed a time course experiment, tracking simultaneously transcript abundance and lipid content of Nannochloropsis cultures grown in N sufficient (+N) and N depleted (-N) media. The data and their analysis were published on Plant Cell (doi: http://dx.doi.org/10.1105/tpc.113.121418). Full reference is reported at the end of this post (reference n 1).
and that are in agreement with the profile of transcript and protein abundance in Nannochloropsis‘ cultures that accumulate lipids. New experimental data became recently available for the scientific community concerning this issue. Jing Li and coworkers performed a time course experiment, tracking simultaneously transcript abundance and lipid content of Nannochloropsis cultures grown in N sufficient (+N) and N depleted (-N) media. The data and their analysis were published on Plant Cell (doi: http://dx.doi.org/10.1105/tpc.113.121418). Full reference is reported at the end of this post (reference n 1).
The glycerolipid profiling performed by Li and coworkers revealed small amounts of TAGs in cells of N. oculata IMET1 grown in Nitrogen sufficient media and the amount of TAGs was found to increase by about 3 folds when the cultures reached the end of the lag phase. In Nitrogen depleted cultures the amount of TAGs increased by approximately 400 folds in 96 hours after Nitrogen depletion and at the same time photosynthetic membrane lipids decreased significantly. Lipase-mediated membrane degradation is likely to channel FA from photosynthetic membranes to TAGs in Nitrogen depletion and indeed 14 of the 34 putative lipases identified in the genome, were up-regulated to some extent in Nitrogen scarcity. As previously observed by Simionato et al. 2013, however, the decrease of membrane lipids is not enough to account for the increase of TAGs and de novo biosynthesis of FA must take place in the cells in Nitrogen scarcity.
According to the new experimental data, the authors hypothesise that most of the proteins involved in de novo synthesis of FA (ACCase; ACP; KAS) are always abundant in the cells, both in +N and in -N conditions, and that the increase in FA must be rather due to an increased flux of precursors that enter the FA‘s biosynthetic pathway. Previous data led other authors to drive similar conclusions (Radakovits et al. 2012; Corteggiani Carpinelli et al. 2013)
Thanks to the new experimental evidence the author argue that the data suggest the presence of multiple sub-cellular compartments involved in FA and TAG biosynthesis and that the vast majority of the newly synthesised FAs are produced in the plastid through the de novo prokaryotic pathway.
The global transcriptomic analysis suggested that various pathways, which are up-regulated in -N, could supply carbon precursors for FA synthesis during Nitrogen starvation:
- the cytosolic glycolysis pathway, which produces pyruvate
- the PDHC bypass, which yields additional acetyl-CoA
- the coupling of TCA reactions with mitochondrial b-oxidation.
The authors observe that the increase of transcripts involved in carbohydrate and protein degradation along with the identification of genes encoding plastid and mitochondrial membrane transporters suggested a shunting of carbon precursors from carbohydrate and protein catabolism into FA synthesis and ultimately TAG synthesis under nitrogen deprivation.
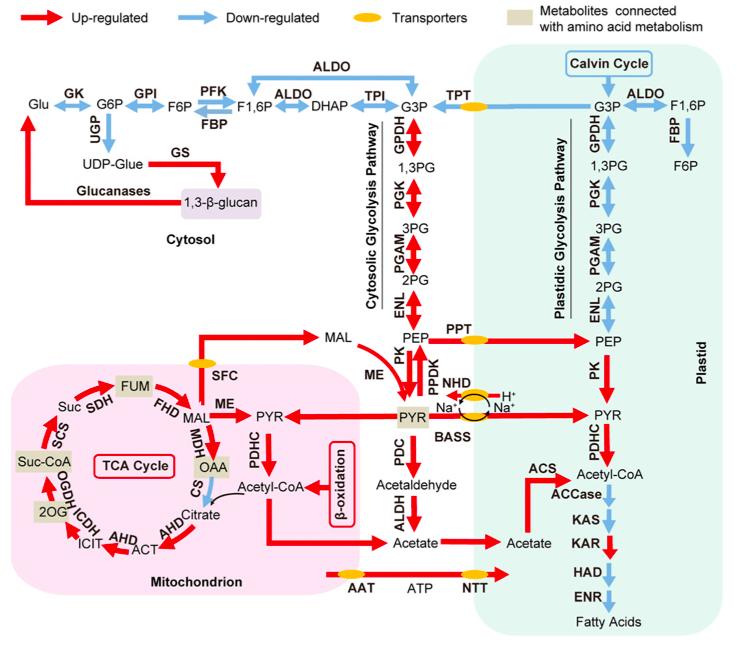
Schematic representation of the Central Carbon Metabolism in N. oculata in Nitrogen depletion from Li J et al. 2014
Finally, however, the authors suggest that, despite the increased production of FA during Nitrogen starvation, the regulatory step that determines the final TAG concentration in -N is actually the TAG assembly pathway. In oder words, under N depletion, it is the up-regulation of the transcript level of the key genes in TAG assembly, rather than those in FA biosynthesis, that leads to accelerated TAG production. Moreover the progressive up-regulation of various DGAT in Nitrogen depletion is likely very important for the bulk of the TAGs formed in -N.
I hope that this post will prompt an active debate on the topic!
References
- Li J, Han D, Wang D, Ning K, Jia J, Wei L, Jing X, Huang S, Chen J, Li Y, Hu Q, Xu J “Choreography of Transcriptomes and Lipidomes of Nannochloropsis Reveals the Mechanisms of Oil Synthesis in Microalgae.”The Plant Cell April 2014. doi: http://dx.doi.org/10.1105/tpc.113.121418
- Simionato D, Block MA, La Rocca N, Jouhet J, Maréchal E, Finazzi G, Morosinotto T. (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganisation of the photosynthetic apparatus. Eukaryotic Cell May 2013 vol. 12 no. 5 665-676
- Radakovits, R. et al. “Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana.” Nat Comms 3, 686 (2012).
- Corteggiani Carpinelli E, Telatin A, Vitulo N, Forcato C, D’Angelo M, Schiavon R, Vezzi A, Giacometti GM, Morosinotto T, Valle G. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120.
- Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR (2013) Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiology, 162:1110–1126
Recent Comments