Proteomics and transcriptomics findings on lipids accumulation in Nannochloropsis

Confocal microscopy image of Nannochloropsis cell grown in nitrogen deprivation. Lipid droplets are in yellow and chloroplast is in red. From “Going ultra deep to unravel the secret recipe of biofuel” by Elisa Corteggiani Carpinelli
What is the mechanism by which Nannochloropsis cells synthesise and accumulate lipids in certain culturing conditions? And moreover is there a key control that we can manipulate to improve lipid yields and overcome the tradeoff between lipid production and growth? This is probably the “holy grail” of Nannochloropsis research. The general feeling is that we do not have an answer yet, but at least we have some clues to start our quest.
Transcriptomic data on Nannochloropsis gene expression during normal growth and nitrogen starvation (which leads to an evident accumulation of oil) were produced by Radakovits et al. (2012), Viler et al. (2012), Liang et al. (2012), Corteggiani Carpinelli at al. (2013). The all of this data show that genes involved in fatty acid and triacylglycerol biosynthesis are alway abundant in the cells (both when nitrogen is abundant and when it is deficient) and their expression is not correlated with the amount of oil accumulated in the cells. Also the expression of the genes involved in lipid degradation does not seem to change drastically, at least not enough to justify the effects. The general conclusion suggested by these experiments is that Nannochloropsis constitutively produces lipids and that the metabolic reorganisation that follows nitrogen deprivation increases the flux of substrates through this pathway, which is in turn capable to sustain the increased metabolic flux.
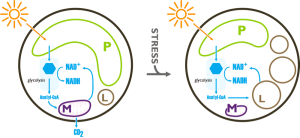
Using the available experimental evidence Corteggiani Carpinelli and coworkers (2013) propose a model of the metabolic response of Nannochloropsis to nitrogen deficiency that can lead to overproduction of fatty acids and as a consequence to the accumulation of oil. They remark that one of the most evident effects of nitrogen starvation is the deescalation of the organelles and therefore it must be taken into account in the elaboration of a model of the metabolic reorganisation of Nannochloropsis. Simionato et al. (2012) showed that despite the decrease of photosynthetic yield, photosynthesis is the only energy supply of the cells in nitrogen starvation. The model therefore states that the energy comes into the system through the photosynthetic activity of the chloroplast in both nitrogen sufficient and nitrogen depleted cultures, and produces glucose. After degradation of the glucose by glycolysis (the genes of this pathway are expressed at similar levels in both the two culturing conditions) the Acetyl-CoA and and the reduced NAD(P)H are mainly reoxidised through the Krebs cycle and the mitochondrial respiration in nitrogen sufficient cultures. Only a small fraction of the Acetyl-CoA and and the reduced NAD(P)H enter the fatty acid biosynthetic pathway in this condition. During nitrogen starvation, due to the severe down-regulation of the mitochondrial genes the reoxidation of the Acetyl-CoA and and the reduced NAD(P)H becomes less efficient and more substrates are available to enter the fatty acid biosynthetic pathway, leading to the accumulation of oil into the lipid droplets.
Potential Model of the flux trough the fatty acid synthesis pathways in nitrogen sufficient and nitrogen deficient culturing conditions based on transcriptomic data, elaborated by Corteggiani Carpinelli and coworkers (2013), drawn by Andrea Telatin. P, plastid; M, mitochondrion; L, lipid droplet.
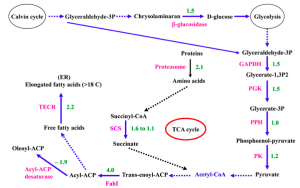
Dong et al. (2013) also examined Nannochloropsis response to nitrogen deficiency and compared protein abundance in parallele cultures grown in nitrogen sufficient and nitrogen deficient media. The experimental conditions of this experiment are different from those used in the various transcriptomic surveys: first of all the cells were supplied with CO2 during growth and moreover the time-course of the nitrogen deprivation experiment was different. Nevertheless also in this proteomic study the data seems to support the hypothesis that the accumulation of triacylglycerols is due to an increase of the metabolic flux trough the fatty acid biosynthetic pathway. The authors report that several key enzymes involved in glycolysis increase during nitrate depletion as well as enzymes involved in fatty acids synthesis and in chrysolaminaran hydrolysation. They therefore advance the hypothesis that, in their experimental conditions, the degradation of storage sugars and the up-regulation of glycolysis are responsible for the increase of substrates through the pathway.
Potential model of fatty acid synthesis pathways based on protein changes during chronic nitrogen depletion, elaborated by Dong and coworkers (2013). SCS, Succinyl-CoA synthetase; TECR, trans-2-enoyl-CoA reductase. The numbers represent fold change of proteins in relative abundance. Dashed lines mean that no related proteins were detected in this pathway.
A summary of the informations obtained studying the accumulation of lipids in Nannochloropsis in response to nitrogen deprivation is also reported on Wikipedia!
References
- Corteggiani Carpinelli, E. et al. “Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion.” Molecular Plant (2014) 7 (2): 323-335. doi: 10.1093/mp/sst120.
- Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR (2013) Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiology, 162:1110–1126
- Liang, C. et al. “De Novo Sequencing and Global Transcriptome Analysis of Nannochloropsis sp. (Eustigmatophyceae) Following Nitrogen Starvation.” Bioenerg. Res. 6, 494–505 (2012).
- Radakovits, R. et al. “Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana.” Nat Comms 3, 686 (2012).
- Simionato D, Block MA, La Rocca N, Jouhet J, Maréchal E, Finazzi G, Morosinotto T. (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganisation of the photosynthetic apparatus. Eukaryotic Cell May 2013 vol. 12 no. 5 665-676
- Vieler, A. et al. “Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga Nannochloropsis oceanica CCMP1779.” PLoS Genet 8, e1003064 (2012).
Share this:
Tags: lipids, metacyc, nannochloropsis rna-seq, nitrogen deprivation, organelles, proteomics, transcriptomics, wikipedia

Recent Comments